Troubleshooting#
Here are some ‘common’ errors that you might encounter at some point:
Issue with X Forwarding#
If you forget to request X forwarding (graphic forwarding) when connecting, you will end up with errors looking like:
qt.qpa.xcb: could not connect to display
qt.qpa.plugin: Could not load the Qt platform plugin "xcb" in "" even though it was found.
This application failed to start because no Qt platform plugin could be initialized. Reinstalling the application may fix this problem.
Available platform plugins are: eglfs, linuxfb, minimal, minimalegl, offscreen, vnc, wayland-egl, wayland, wayland-xcomposite-egl, wayland-xcomposite-glx, xcb.
Aborted (core dumped)
As a reminder to connect:
To ‘slurm-cluster’:
ssh -XC {user}@cluster-access
When using salloc with SLURM, use the -x11 option like:
salloc --partition gpu --x11 --gres=gpu:1 --mem=256G srun --pty bash -l
Issues with Qt (Application Crashes with Segfault)#
The Tomwer GUI uses silx from the silx.gui.qt module, which tries to use PyQt5, then PySide2, then PyQt4. The order is different for AnyQt, used by Orange3. This discrepancy might lead to errors in Signal/SLOT connections during widget instantiation.
Be careful if more than one Qt binding is available. A quick workaround is to change the AnyQt/__init__.py file (e.g., the ‘availableapi’ function).
Issue with Silx Library and Static TLS#
It is known that with some OS (e.g., Debian 8) we can encounter issues with OpenMP. Usually, we compile silx with the –no-openmp flag (as the distributed wheel). Otherwise, errors like [this issue](silx-kit/silx#3102) can occur.
At Tomwer’s side, the only usage of OpenMP is with the median filter, so it’s not considered a major issue if OpenMP is deactivated.
Issue with Reconstruction Being Slow#
If reconstruction on a GPU appears to be very slow (several minutes), the issue might be that the scan dataset is stored in several files. For example, in HDF5 files, if each frame is stored in a dedicated file.
Issue with the ‘datalistener’#
If the data listener is started and no acquisition/scan is found, there are two possible issues:
The tomo-sync from Bliss-tomo is not activated. You can check this from bliss multivisor.
The configuration of Bliss-tomo is not sending the RPC command to the computer running Tomwer. As the configuration seems to evolve and there is no fixed rule, please check with BCU.
Additionally, the latest data-listener now listens on the ‘targeted port’ by default. Previously (version < 1.0), users had to do this manually using the tomwer stop-data-listener command. This can still fail if the user launching Tomwer does not have the right to stop the ‘owning port’ process.
Slice Not Being Reconstructed / Taking Much Time#
A common issue is that the slice is not reconstructed or takes much time (in recent Nabu versions > 2023) when the user tries to reconstruct a slice without having a GPU.
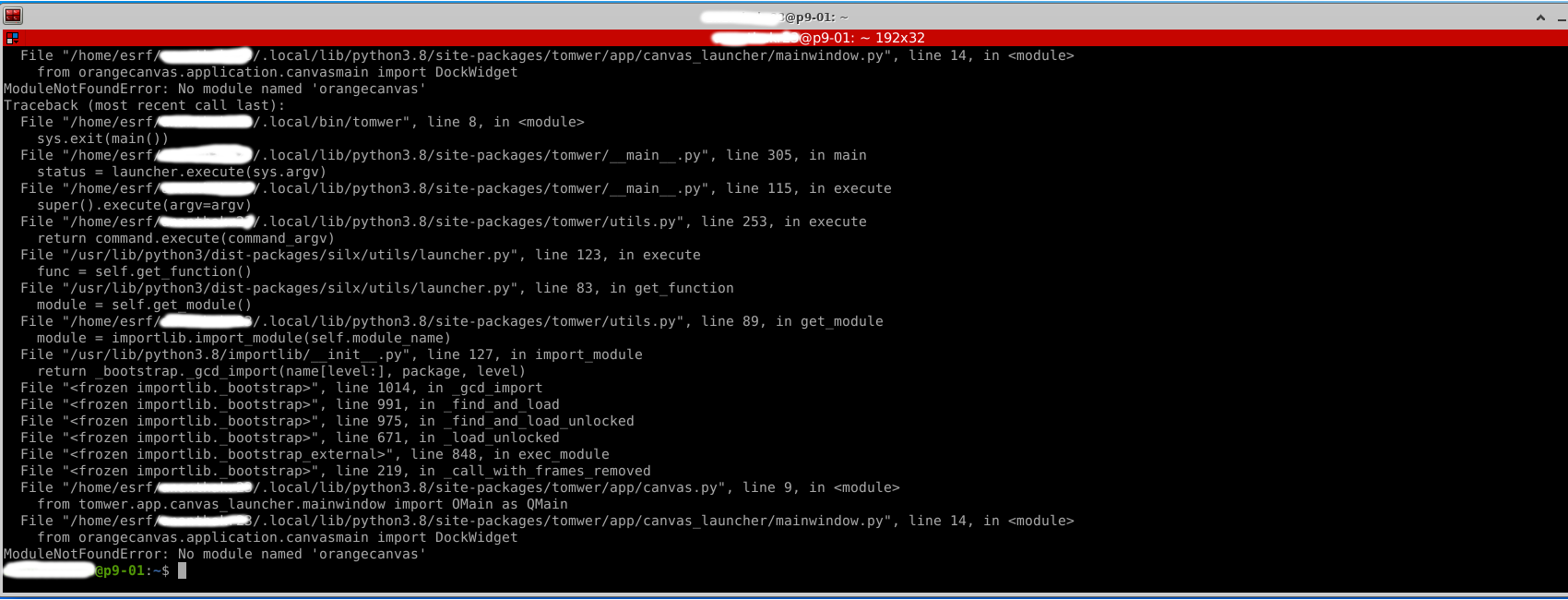
Issue with Missing Python Library#
It can happen that a user has installed Tomwer locally and then executes module load tomotools. This will trigger the local installation. If the local installation was done without the pip install tomwer [full] command, you might encounter errors like:
Workflow processing#
Issue with a Widget Processing the Same Dataset Multiple Times#
We have encountered issues where a widget sends the same ‘data’ or ‘volume’ multiple times during processing, triggering downstream processing more than once.
This is clearly a bug. If you encounter it, please contact us and/or create an issue about it: https://gitlab.esrf.fr/tomotools/tomwer/-/issues
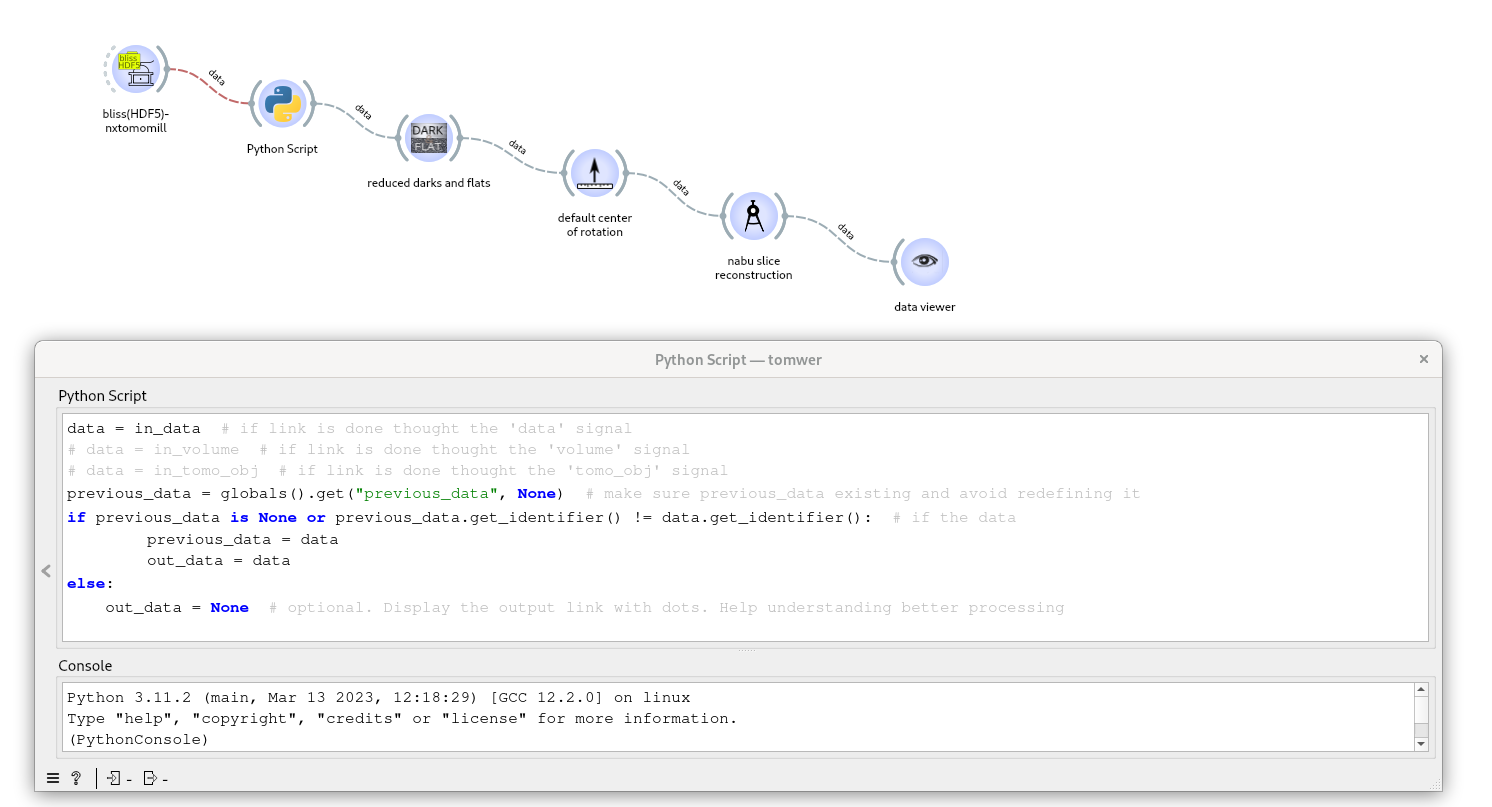
One way to bypass this issue is to add a small Python script after the widget that emits the signal multiple times. This script prevents sending the same signal twice in a row:
data = in_data # if link is done through the 'data' signal
# data = in_volume # if link is done through the 'volume' signal
# data = in_tomo_obj # if link is done through the 'tomo_obj' signal
previous_data = globals().get("previous_data", None) # make sure previous_data exists and avoid redefining it
if previous_data is None or previous_data.get_identifier() != data.get_identifier(): # if the data has not been sent last iteration
previous_data = data
out_data = data
else:
out_data = None # optional. Display the output link with dots. Helps understand processing better
For example, if the “Bliss (HDF5) - Nxtomomill” sends a dataset to be filtered twice, you can add this script to the workflow as shown below:
Workflow processing#