Ewoks Tutorial 1 - Launching a Workflow Using ewoks execute#
Since version 1.1, you can execute and convert Orange workflow files (.ows) using ewoks.
Requirements#
Have a tomography data processing workflow already defined and saved as .ows. (processing_data.ows in this example).
Tomwer >= 1.1 installed.
Execute the Workflow from the Command Line Interface#
We will use ewoks execute and provide some input(s).
Currently, users can provide at most one scan or one volume.
For this tutorial, we will provide a scan (NXtomo) to the processing_data.ows file workflow. It will define the data input in the Tomwer workflow.
Let’s assume the NXtomo is saved in ‘my_dataset.nx’ under the entry HDF5 group. The identifier for this example is:
hdf5:scan:my_dataset.nx?path=entry0000
See the identifiers definition from tomoscan for more information.
To execute the workflow with ewoks, run:
ewoks execute processing_data.json --parameter data=hdf5:scan:my_dataset.nx?path=entry0000
If providing a volume (e.g., for a workflow with volume casting), the URL would look like:
For a volume stored in HDF5:
hdf5:volume:./bambou_hercules_0001slice_1080.hdf5?path=entry0000/reconstructionFor a volume stored in TIFF (one frame per file):
tiff:volume:/path/to/my/my_folder
For a volume stored in EDF:
edf:volume:/path/to/my/my_folder
For a volume stored in JPEG 2000:
jp2k:volume:/path/to/my/my_folder
The execution of the workflow would look like this:
ewoks execute processing_volume.ows --parameter volume={volume_url}
Ewoks Python API#
You can also use the Ewoks Python API to execute workflows directly from Ewoks. For details, please refer to the ewoks python API.
Limitations#
Python Script#
For now, Python scripts from Orange Canvas only handle ‘data’ and ‘volume’ inputs/outputs when executed with ewoks.
Convert from Orange Workflow (.ows) to Ewoks Workflow#
For this example, we will consider the following workflow saved under the processing_data.ows file:
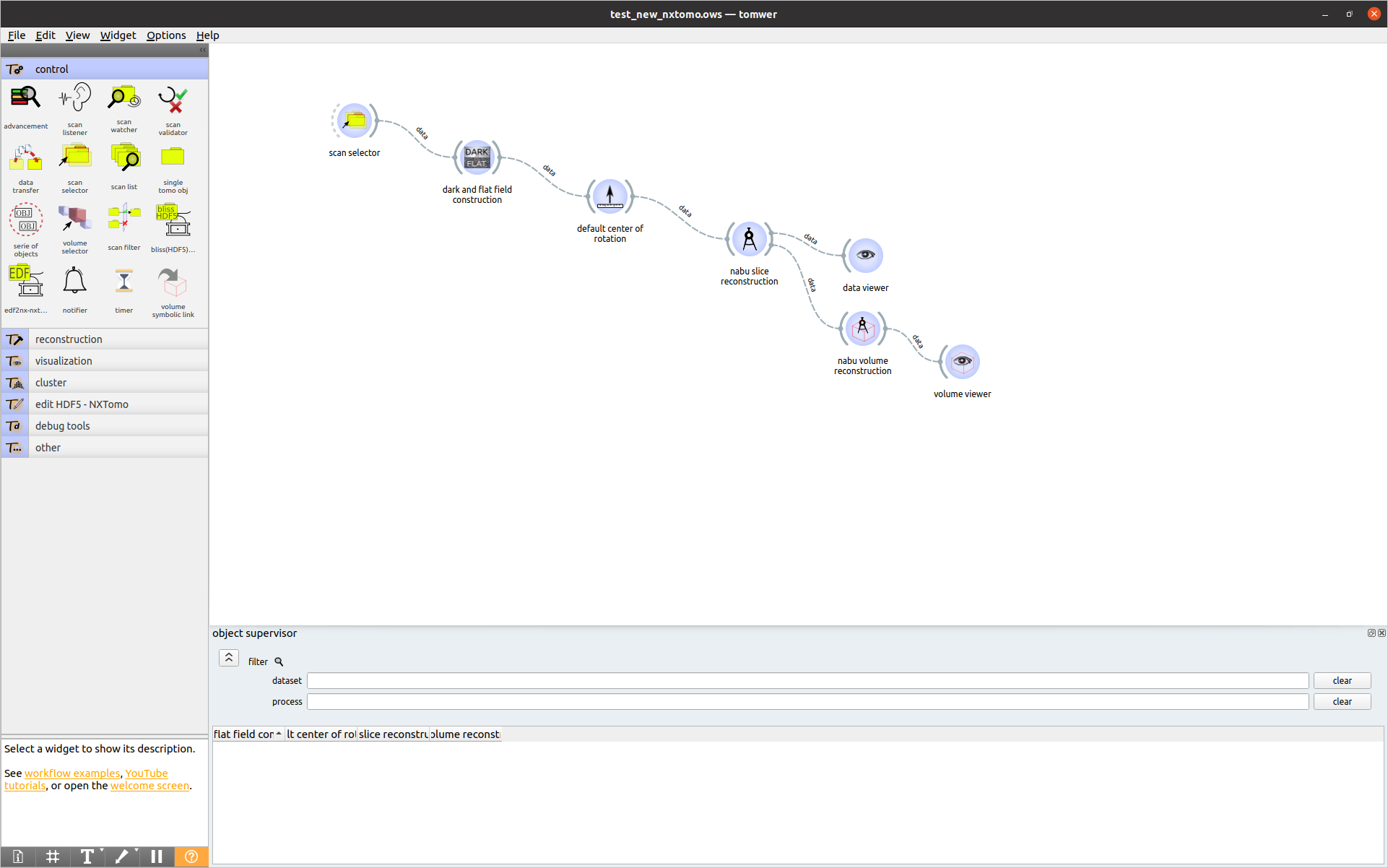
The .ows file looks like:
<?xml version='1.0' encoding='utf-8'?>
<scheme version="2.0" title="" description="">
<nodes>
<node id="0" name="scan selector" qualified_name="orangecontrib.tomwer.widgets.control.DataSelectorOW.DataSelectorOW" project_name="tomwer" version="" title="scan selector" position="(205.0, 97.0)" />
<node id="1" name="dark and flat field construction" qualified_name="orangecontrib.tomwer.widgets.reconstruction.DarkRefAndCopyOW.DarkRefAndCopyOW" project_name="tomwer" version="" title="dark and flat field construction" position="(361.0, 156.0)" />
<node id="2" name="default center of rotation" qualified_name="orangecontrib.tomwer.widgets.reconstruction.AxisOW.AxisOW" project_name="tomwer" version="" title="default center of rotation" position="(520.0, 207.0)" />
<node id="3" name="nabu slice reconstruction" qualified_name="orangecontrib.tomwer.widgets.reconstruction.NabuOW.NabuOW" project_name="tomwer" version="" title="nabu slice reconstruction" position="(683.0, 246.0)" />
<node id="4" name="nabu volume reconstruction" qualified_name="orangecontrib.tomwer.widgets.reconstruction.NabuVolumeOW.NabuVolumeOW" project_name="tomwer" version="" title="nabu volume reconstruction" position="(840.0, 370.0)" />
<node id="5" name="volume viewer" qualified_name="orangecontrib.tomwer.widgets.visualization.VolumeViewerOW.VolumeViewerOW" project_name="tomwer" version="" title="volume viewer" position="(987.0, 440.0)" />
<node id="6" name="data viewer" qualified_name="orangecontrib.tomwer.widgets.visualization.DataViewerOW.DataViewerOW" project_name="tomwer" version="" title="data viewer" position="(848.0, 242.0)" />
</nodes>
<links>
<link id="0" source_node_id="0" sink_node_id="1" source_channel="data" sink_channel="data" enabled="true" />
<link id="1" source_node_id="1" sink_node_id="2" source_channel="data" sink_channel="data" enabled="true" />
<link id="2" source_node_id="2" sink_node_id="3" source_channel="data" sink_channel="data" enabled="true" />
<link id="3" source_node_id="3" sink_node_id="4" source_channel="data" sink_channel="data" enabled="true" />
<link id="4" source_node_id="4" sink_node_id="5" source_channel="data" sink_channel="data" enabled="true" />
<link id="5" source_node_id="3" sink_node_id="6" source_channel="data" sink_channel="data" enabled="true" />
</links>
<annotations />
<thumbnail />
<node_properties>
<properties node_id="0" format="literal">{'_scanIDs': [], 'controlAreaVisible': True, 'savedWidgetGeometry': None, '__version__': 1}</properties>
<properties node_id="1" format="literal">{'_ewoks_default_inputs': {'data': None, 'dark_ref_params': None}, '_rpSetting': {}, 'controlAreaVisible': True, 'savedWidgetGeometry': None, '__version__': 1}</properties>
<properties node_id="2" format="literal">{'_ewoks_default_inputs': {'data': None, 'axis_params': {'MODE': 'sino-coarse-to-fine', 'POSITION_VALUE': None, 'CALC_INPUT_TYPE': 'transmission_nopag', 'ANGLE_MODE': '0-180', 'USE_SINOGRAM': True, 'SINOGRAM_LINE': 'middle', 'SINOGRAM_SUBSAMPLING': 10, 'AXIS_URL_1': '', 'AXIS_URL_2': '', 'LOOK_AT_STDMAX': False, 'NEAR_WX': 5, 'FINE_STEP_X': 0.1, 'SCALE_IMG2_TO_IMG1': False, 'NEAR_POSITION': 0.0, 'PADDING_MODE': 'edge', 'FLIP_LR': True, 'COMPOSITE_OPTS': {'theta': 10, 'oversampling': 4, 'n_subsampling_y': 10, 'take_log': True, 'near_pos': 0.0, 'near_width': 20}, 'SIDE': 'left', 'COR_OPTIONS': ''}, 'gui': {'mode_is_lock': True, 'value_is_lock': False, 'auto_update_estimated_cor': True}}, '_rpSetting': {}, 'controlAreaVisible': True, 'savedWidgetGeometry': b'\x01\xd9\xd0\xcb\x00\x03\x00\x00\x00\x00\x01}\x00\x00\x00\xe2\x00\x00\x06\x03\x00\x00\x03\xa9\x00\x00\x01}\x00\x00\x01\x07\x00\x00\x06\x03\x00\x00\x03\xa9\x00\x00\x00\x00\x00\x00\x00\x00\x07\x80\x00\x00\x01}\x00\x00\x01\x07\x00\x00\x06\x03\x00\x00\x03\xa9', '__version__': 1}</properties>
<properties node_id="3" format="literal">{'_ewoks_default_inputs': {'data': None, 'nabu_params': {'preproc': {'flatfield': 1, 'double_flatfield_enabled': 0, 'dff_sigma': 0.0, 'ccd_filter_enabled': 0, 'ccd_filter_threshold': 0.04, 'take_logarithm': True, 'log_min_clip': 1e-06, 'log_max_clip': 10.0, 'sino_rings_correction': 'None', 'sino_rings_options': 'sigma=1.0 ; levels=10', 'tilt_correction': '', 'autotilt_options': ''}, 'reconstruction': {'method': 'FBP', 'angles_file': '', 'axis_correction_file': '', 'angle_offset': 0.0, 'fbp_filter_type': 'ramlak', 'padding_type': 'edges', 'iterations': 200, 'optim_algorithm': 'chambolle-pock', 'weight_tv': 0.01, 'preconditioning_filter': 1, 'positivity_constraint': 1, 'rotation_axis_position': '', 'translation_movements_file': '', 'clip_outer_circle': 1, 'centered_axis': 1, 'start_x': 0, 'end_x': -1, 'start_y': 0, 'end_y': -1, 'start_z': 0, 'end_z': -1, 'enable_halftomo': 0}, 'dataset': {'binning': 1, 'binning_z': 1, 'projections_subsampling': 1}, 'tomwer_slices': 'middle', 'output': {'file_format': 'hdf5', 'location': ''}, 'phase': {'method': 'Paganin', 'delta_beta': '100.0', 'padding_type': 'edge', 'unsharp_coeff': 0, 'unsharp_sigma': 0, 'ctf_geometry': ' z1_v=None; z1_h=None; detec_pixel_size=None; magnification=True', 'beam_shape': 'parallel', 'ctf_advanced_params': ' length_scale=1e-05; lim1=1e-05; lim2=0.2; normalize_by_mean=True', 'ctf_translations_file': ''}, 'configuration_level': 'optional', 'mode_locked': False, 'cluster_config': None}}, '_rpSetting': {'preproc': {'flatfield': 1, 'double_flatfield_enabled': 0, 'dff_sigma': 0.0, 'ccd_filter_enabled': 0, 'ccd_filter_threshold': 0.04, 'take_logarithm': True, 'log_min_clip': 1e-06, 'log_max_clip': 10.0, 'sino_rings_correction': 'None', 'sino_rings_options': 'sigma=1.0 ; levels=10', 'tilt_correction': '', 'autotilt_options': ''}, 'reconstruction': {'method': 'FBP', 'angles_file': '', 'axis_correction_file': '', 'angle_offset': 0.0, 'fbp_filter_type': 'ramlak', 'padding_type': 'edges', 'iterations': 200, 'optim_algorithm': 'chambolle-pock', 'weight_tv': 0.01, 'preconditioning_filter': 1, 'positivity_constraint': 1, 'rotation_axis_position': '', 'translation_movements_file': '', 'clip_outer_circle': 1, 'centered_axis': 1, 'start_x': 0, 'end_x': -1, 'start_y': 0, 'end_y': -1, 'start_z': 0, 'end_z': -1, 'enable_halftomo': 0}, 'dataset': {'binning': 1, 'binning_z': 1, 'projections_subsampling': 1}, 'tomwer_slices': 'middle', 'output': {'file_format': 'hdf5', 'location': ''}, 'phase': {'method': 'Paganin', 'delta_beta': '100.0', 'padding_type': 'edge', 'unsharp_coeff': 0, 'unsharp_sigma': 0, 'ctf_geometry': ' z1_v=None; z1_h=None; detec_pixel_size=None; magnification=True', 'beam_shape': 'parallel', 'ctf_advanced_params': ' length_scale=1e-05; lim1=1e-05; lim2=0.2; normalize_by_mean=True', 'ctf_translations_file': ''}, 'configuration_level': 'optional', 'mode_locked': False, 'cluster_config': None}, 'controlAreaVisible': True, 'savedWidgetGeometry': b'\x01\xd9\xd0\xcb\x00\x03\x00\x00\x00\x00\x02\x03\x00\x00\x014\x00\x00\x05|\x00\x00\x03{\x00\x00\x02\x03\x00\x00\x014\x00\x00\x05|\x00\x00\x03{\x00\x00\x00\x00\x00\x00\x00\x00\x07\x80\x00\x00\x02\x03\x00\x00\x014\x00\x00\x05|\x00\x00\x03{', '__version__': 1}</properties>
<properties node_id="4" format="literal">{'_ewoks_default_inputs': {'data': None, 'nabu_volume_params': None, 'nabu_params': None}, '_rpSetting': {'start_z': 30, 'end_z': 35, 'gpu_mem_fraction': 0.9, 'cpu_mem_fraction': 0.9, 'use_phase_margin': True, 'postproc': {'output_histogram': 1}, 'new_output_file_format': '', 'new_output_location': '', 'cluster_config': None}, 'controlAreaVisible': True, 'savedWidgetGeometry': b'\x01\xd9\xd0\xcb\x00\x03\x00\x00\x00\x00\x02\xd5\x00\x00\x01\xbc\x00\x00\x04\xaa\x00\x00\x02\xf3\x00\x00\x02\xd5\x00\x00\x01\xbc\x00\x00\x04\xaa\x00\x00\x02\xf3\x00\x00\x00\x00\x00\x00\x00\x00\x07\x80\x00\x00\x02\xd5\x00\x00\x01\xbc\x00\x00\x04\xaa\x00\x00\x02\xf3', '__version__': 1}</properties>
<properties node_id="5" format="literal">{'controlAreaVisible': True, 'savedWidgetGeometry': None, '__version__': 1}</properties>
<properties node_id="6" format="literal">{'_viewer_config': {}, 'controlAreaVisible': True, 'savedWidgetGeometry': None, '__version__': 1}</properties>
</node_properties>
<session_state>
<window_groups />
</session_state>
</scheme>
Now we can convert this file to an Ewoks workflow file using the ewoks convert command. In this case, we will choose JSON as the format:
ewoks convert processing_data.ows processing_data.json
You can open the .json file. The settings of the different tasks are defined by the ‘default_inputs’ keys, such as:
{
"directed": true,
"multigraph": false,
"graph": {
"id": "processing_data",
"label": "Ewoks workflow 'processing_data'",
"schema_version": "1.0"
},
"nodes": [
...
{
...
"default_inputs": [
{
"name": "axis_params",
"value": {
"MODE": "sino-coarse-to-fine",
"POSITION_VALUE": null,
"CALC_INPUT_TYPE": "transmission_nopag",
"ANGLE_MODE": "0-180",
"USE_SINOGRAM": true,
"SINOGRAM_LINE": "middle",
"SINOGRAM_SUBSAMPLING": 10,
"NEAR_WX": 5,
"FINE_STEP_X": 0.1,
"SCALE_IMG2_TO_IMG1": false,
"NEAR_POSITION": 0.0,
"PADDING_MODE": "edge",
"COMPOSITE_OPTS": {
"theta": 10,
"oversampling": 4,
"n_subsampling_y": 10,
"take_log": true,
"near_pos": 0.0,
"near_width": 20
},
"SIDE": "left",
"COR_OPTIONS": ""
}
},
],
"id": "2"
},
...
],
"links": [
...
]
}
You can then execute the .json file the same way you would execute the .ows file. See Execute the Workflow from the Command Line Interface.
Note
Widgets that do not perform any processing (such as pure visualization widgets) will be converted to an ‘ewoks neutral task,’ simply passing the dataset or volume to the next task when necessary. Otherwise, they will execute the run method of the Ewoks Task class. Each Tomwer task implements this function. Task classes are stored in tomwer.core.process.