HDF5Converter#
The more it goes the more use cases the hdf5 converter has to handle. Today it handles “classical” tomography acquisition, zseries and pcotomo.
Behavior of the HDF5Converter#
The HDF5 is applying sequentially the following taks:
preprocess raw data (During converter creation)
browse raw data and collect information on Bliss scan sequence.
During this step it will determine for each entry if the frame type are dark, flats, projections or alignment.
Create an instance of Acquistion for each ‘init’ Bliss scan.
HDF5Converter.convert function
2.1 create for Acquisition instance one or several instances of
NXtomo.2.2 Optional split of NXtomo (use case of pcotomo) in order to obtain the final NXtomo.
2.3 save data to disk.
Acquistion classes#
To manage all the use cases it has evolved. Each acquisition has now it’s own acquisition class. All based on the nxtomomill.converter.hdf5.acquisition.baseacquisition.BaseAcquisition
StandardAcquisitiondefault acquisition.ZSeriesBaseAcquisitiona serie of default acquisition with a set of z values. Create one NXTomo per different z
The regular conversion sequence is:
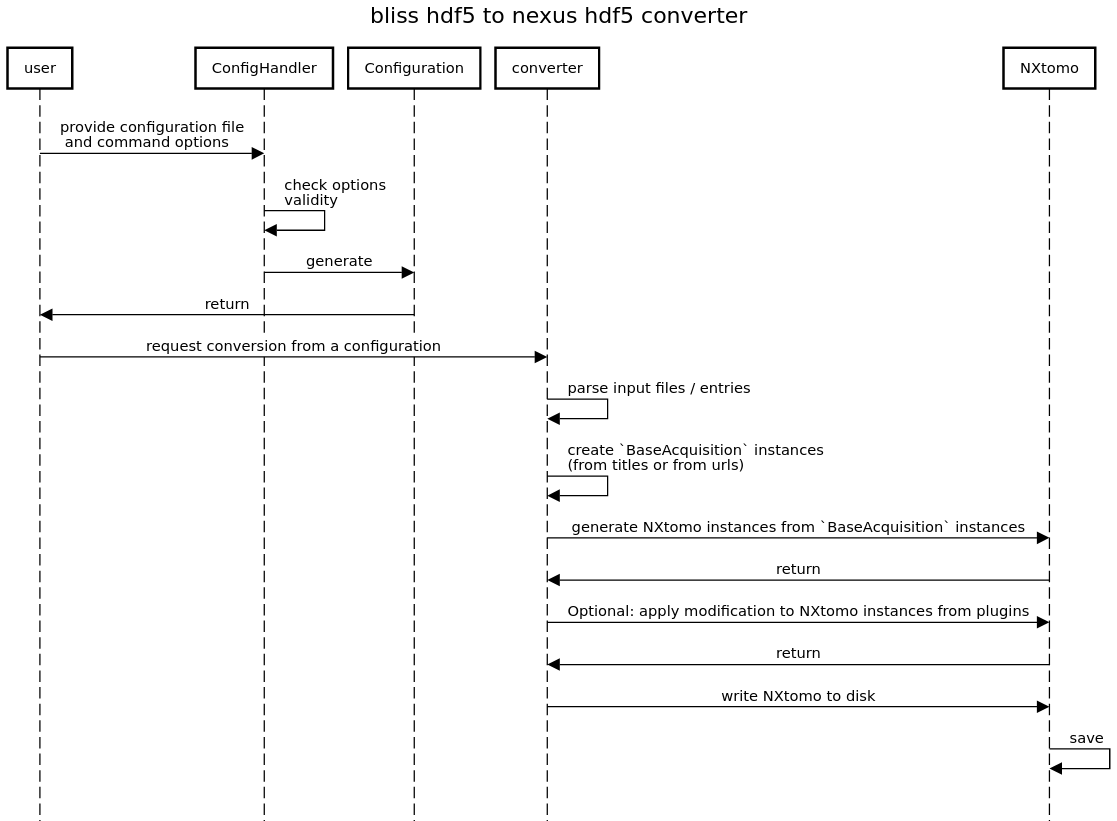
Actually we also have two configuration handler classes. Both inherit from nxtomomill.io.confighandler.BaseHDF5ConfigHandler.
The configuration handler is used to make the connection between applications options and configuration (nxtomomill.io.config.TomoHDF5Config)
TomoHDF5ConfigHandler: used by h52nx application
The configuration is used to make the conversion (defines input file, output file, entry titles, urls to be converted, source format…)
Actually there is also two configuration classes.
TomoHDF5Config: define the configuration to generate a usual “NXTomo”